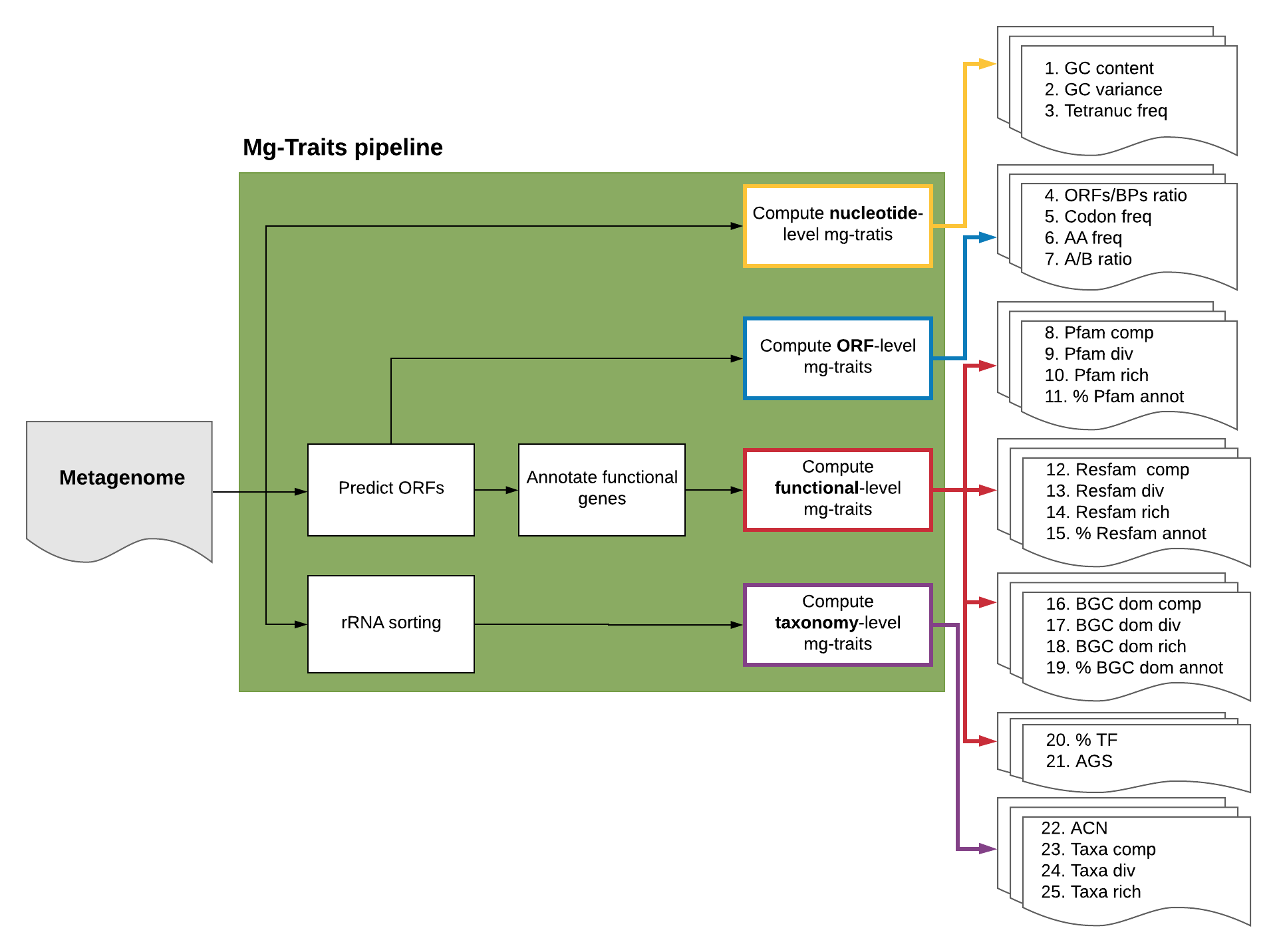
Mg-Traits pipeline. The 25 metagenomic traits computed by the Mg-Traits pipeline are divided into four different groups. The first includes the metagenomic traits computed at the nucleotide level: (1) GC content, (2) GC variance, and (3) Tetranucleotide frequency. The second group includes the traits obtained from the open reading frame (ORF) sequence data: (4) ORFs to Base Pairs (BPs) ratio, (5) Codon frequency, (6) Amino acid frequency, and (7) Acidic to basic amino acid ratio. The third group is based on the functional annotation of the ORF amino acid sequences. The first 12 metagenomic traits (from 8 to 19 in the figure) comprise the composition, diversity, richness, and percentage of annotated genes for three different sets of genes: Pfam (https://pfam.xfam.org), Resfam (http://www.dantaslab.org/resfams), and Biosynthetic Gene Cluster (BGC) domains (https://doi.org/10.1101/2021.01.20.427441). Additionally, this group includes (20) the percentage of transcription factors (TFs) and (21) the average genome size (AGS). Lastly, in the fourth group are included the taxonomy-related metagenomic traits: (22) average copy number of 16S rRNA genes (ACN), taxonomic (23) composition, (24) diversity, and (25) richness.